1.4Network contextualization with differential proteomics data
How to apply a basic network contextualization method to differential proteomics data
1.4.1About¶
This notebook shows how to apply a basic network contextualization method that combines a template prior knowledge network obtained from OmniPath with differential proteomics data from the CPTAC. Specifically, we employ a dataset that characterizes the basal proteomic profiles of lung adenocarcinoma patients (LUAD). The method used in the tutorial is one of the simplest optimization based methods: The Prize-Collecting Steiner Tree, as implemented in CORNETO.
First, we need to install the required packages and load the libraries used in this tutorial:
! pip uninstall -y numpy
! pip install numpy==1.26.41.4.1.1Please restart your session: Runtime -> Restart session¶
# version should be < 2 for pcst_fast compatibility
import numpy as np
print(np.__version__)2.2.5
! pip install pcst_fast
! pip install --upgrade seaborn matplotlib pandas networkximport gdown
import os
# File paths and their corresponding Google Drive IDs
files_to_download = {
"op_signor.csv": "1Ooq4gUlpwZYvUOT3OEueJrqKQyZbxQly",
"luad_de.csv": "1naFI56Wc2glkIn6CG4HJMGRwddxZacME"
}
# Download only if file doesn't exist
for filename, file_id in files_to_download.items():
if not os.path.exists(filename):
print(f"⬇️ Downloading {filename}...")
gdown.download(id=file_id, output=filename)
else:
print(f"✅ {filename} already exists. Skipping download.")import seaborn as sns
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
from pcst_fast import pcst_fast
import networkx as nxAs in previous tutorials, we begin by preparing knowledge component of this analysis. For this network contextualization exercise, we will employ data from Signor, a very curated database of signaling interactions.
signor = pd.read_csv('op_signor.csv')
signor = nx.from_pandas_edgelist(signor[['source_genesymbol', 'target_genesymbol']].rename(columns={'source_genesymbol': 'source', 'target_genesymbol': 'target'}))
signor.number_of_edges(), signor.number_of_nodes()(63683, 6679)We also load and make a first exploration of the differential abundance data.
luad = pd.read_csv('luad_de.csv')
luad['significance'] = -np.log10(luad['adj.P.Val'])
sns.scatterplot(data = luad, x = 'logFC', y ='significance')<Axes: xlabel='logFC', ylabel='significance'>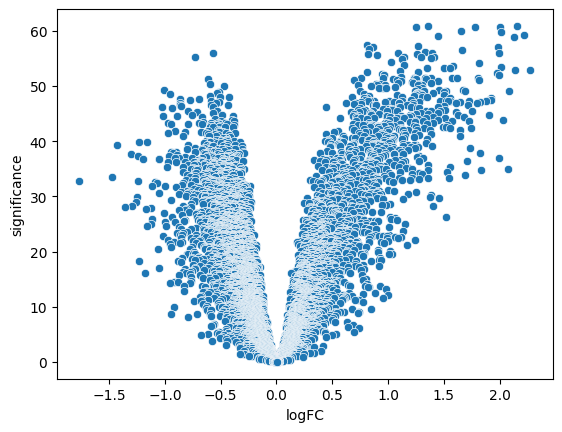
luad.sort_values(by='significance', ascending=False, inplace=True)
luad.head(10)# subset graph to nodes in the luad
signor_luad = signor.subgraph(luad['protein']).copy()
abs_fc_dict = dict(zip(luad['protein'], np.abs(luad['logFC'])))
nx.set_node_attributes(signor_luad, abs_fc_dict, 'logFC')
signor_luad.number_of_edges(), signor_luad.number_of_nodes()(4728, 2747)def solve_pcst(
G: nx.Graph,
prize_attr: str = "logFC",
edge_cost: float = 1.0,
pruning: str = "strong",
verbosity: int = 0,
num_clusters: int = 1
) -> nx.Graph:
"""
Solves a PCST using node prizes and a fixed edge cost (regularization).
Args:
G (nx.Graph): Graph with node attribute `prize_attr`.
prize_attr (str): Node attribute key to use as prize value.
edge_cost (float): Constant edge cost for all edges.
pruning (str): Pruning strategy ('none', 'simple', 'gw', or 'strong').
verbosity (int): Verbosity level (0 for silent, 1 for verbose).
num_clusters (int): Number of desired output components.
Returns:
nx.Graph: Subgraph induced by the PCST solution.
"""
node_list = list(G.nodes())
node_to_idx = {node: i for i, node in enumerate(node_list)}
idx_to_node = {i: node for node, i in node_to_idx.items()}
edge_list = []
cost_list = []
edge_lookup = []
for u, v in G.edges():
edge_list.append([node_to_idx[u], node_to_idx[v]])
cost_list.append(edge_cost)
edge_lookup.append((u, v))
edge_array = np.array(edge_list, dtype=np.int64)
cost_array = np.full(len(edge_list), edge_cost, dtype=np.float64)
prizes = np.array([
G.nodes[node].get(prize_attr, 0.0) for node in node_list
], dtype=np.float64)
prizes = np.abs(prizes)
root_idx = -1
selected_nodes_idx, selected_edges_idx = pcst_fast(
edge_array, prizes, cost_array, root_idx, num_clusters, pruning, verbosity
)
selected_nodes = {idx_to_node[i] for i in selected_nodes_idx}
selected_edges = []
for edge_idx in selected_edges_idx:
u, v = edge_lookup[edge_idx]
if u in selected_nodes and v in selected_nodes:
selected_edges.append((u, v))
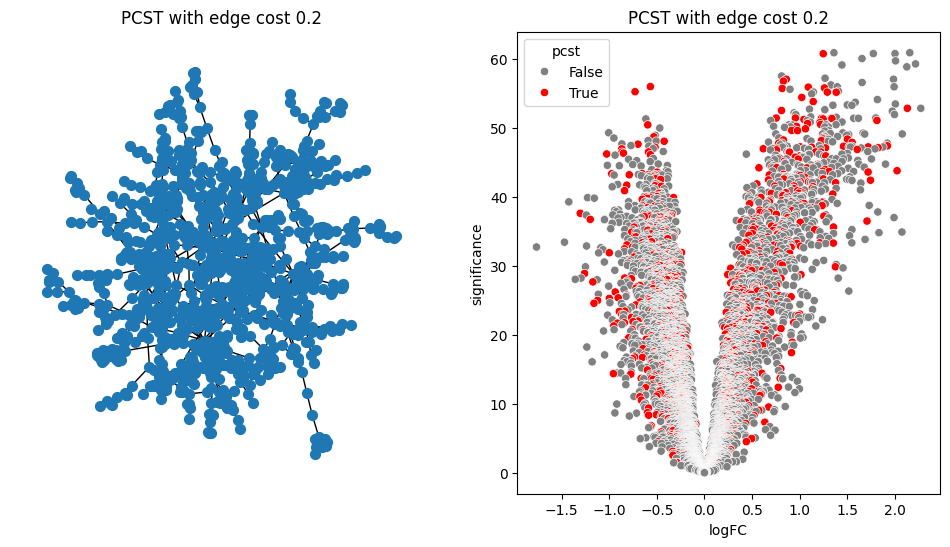
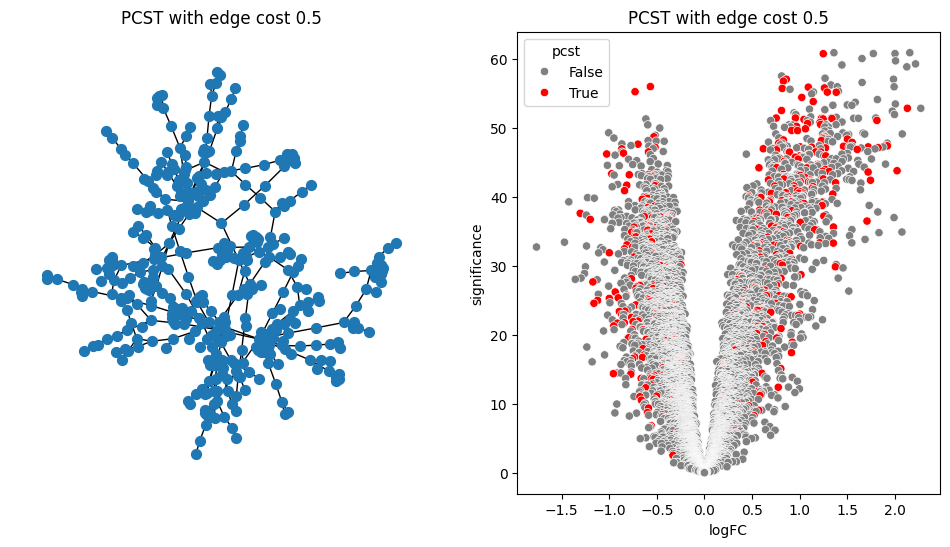
return G.edge_subgraph(selected_edges).copy()for i in [0.2, 0.5, 0.9, 1.0, 1.5]:
pcst = solve_pcst(signor_luad, edge_cost=i)
fig, axs = plt.subplots(1, 2, figsize=(12, 6))
pos = nx.spring_layout(pcst)
lab = len(pcst.nodes()) <= 100
nx.draw(pcst, pos, with_labels=lab, node_size=50, ax=axs[0])
luad['pcst'] = luad['protein'].isin(pcst.nodes())
sns.scatterplot(data=luad, x='logFC', y='significance', hue='pcst', palette=['gray', 'red'], ax=axs[1])
axs[0].set_title(f'PCST with edge cost {i}')
axs[1].set_title(f'PCST with edge cost {i}')/var/folders/bk/4tmncsh16yb13svxsx6w3s2m0000gp/T/ipykernel_61248/2871997957.py:8: UserWarning: The palette list has more values (2) than needed (1), which may not be intended.
sns.scatterplot(data=luad, x='logFC', y='significance', hue='pcst', palette=['gray', 'red'], ax=axs[1])
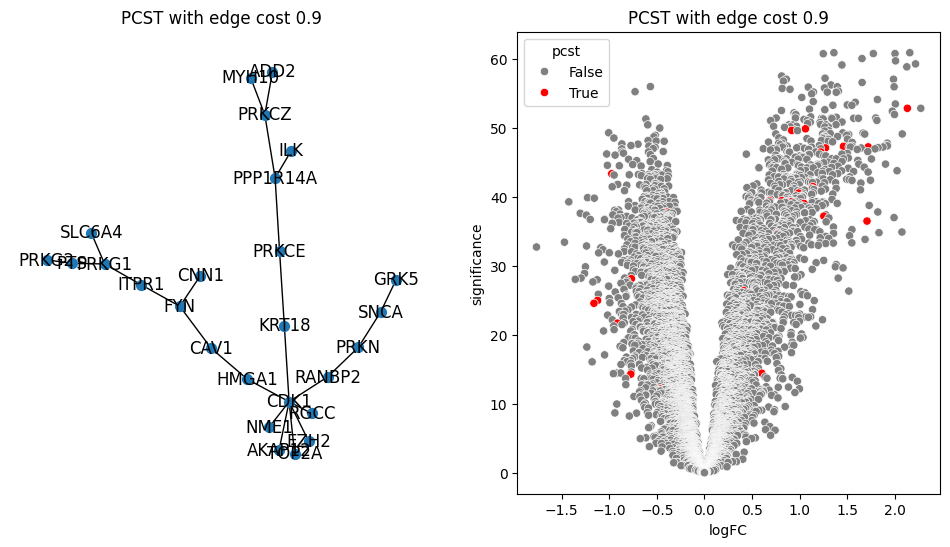
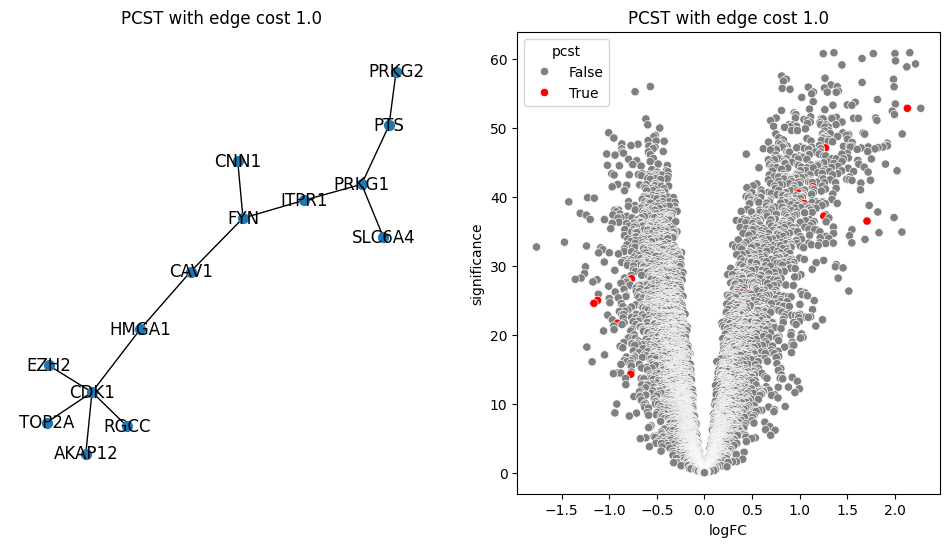
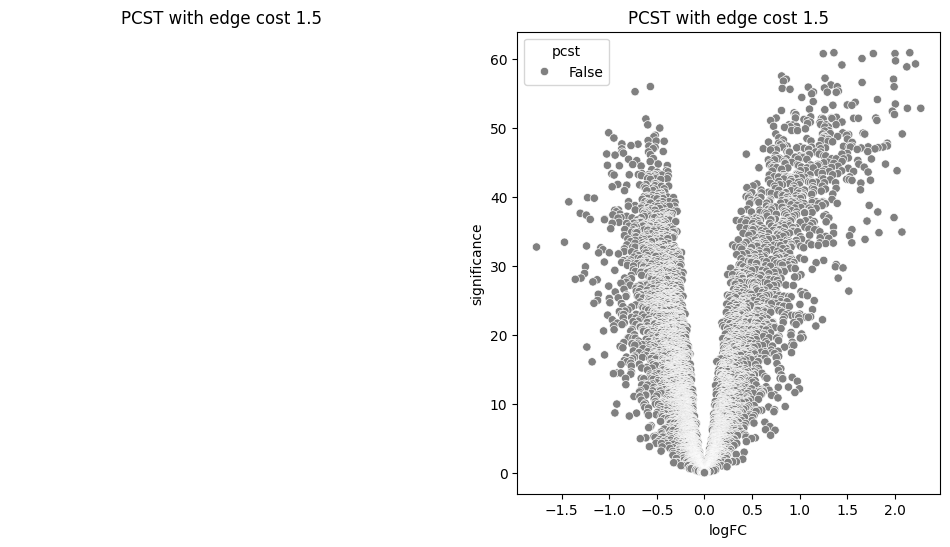
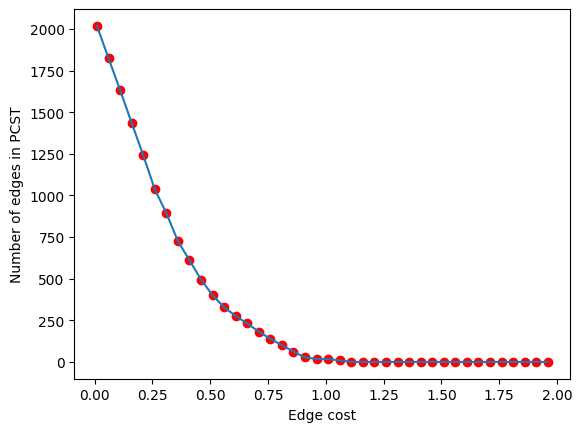
lambdas = np.arange(0.01, 2, 0.05)
pcst_size = []
for i in lambdas:
pcst = solve_pcst(signor_luad, edge_cost=i)
pcst_size.append(len(pcst.edges()))
plt.plot(lambdas, pcst_size)
plt.xlabel('Edge cost')
plt.ylabel('Number of edges in PCST')
plt.scatter(lambdas, pcst_size, color='red')